
Original Papers
- Kengkanna A, Ohue M.
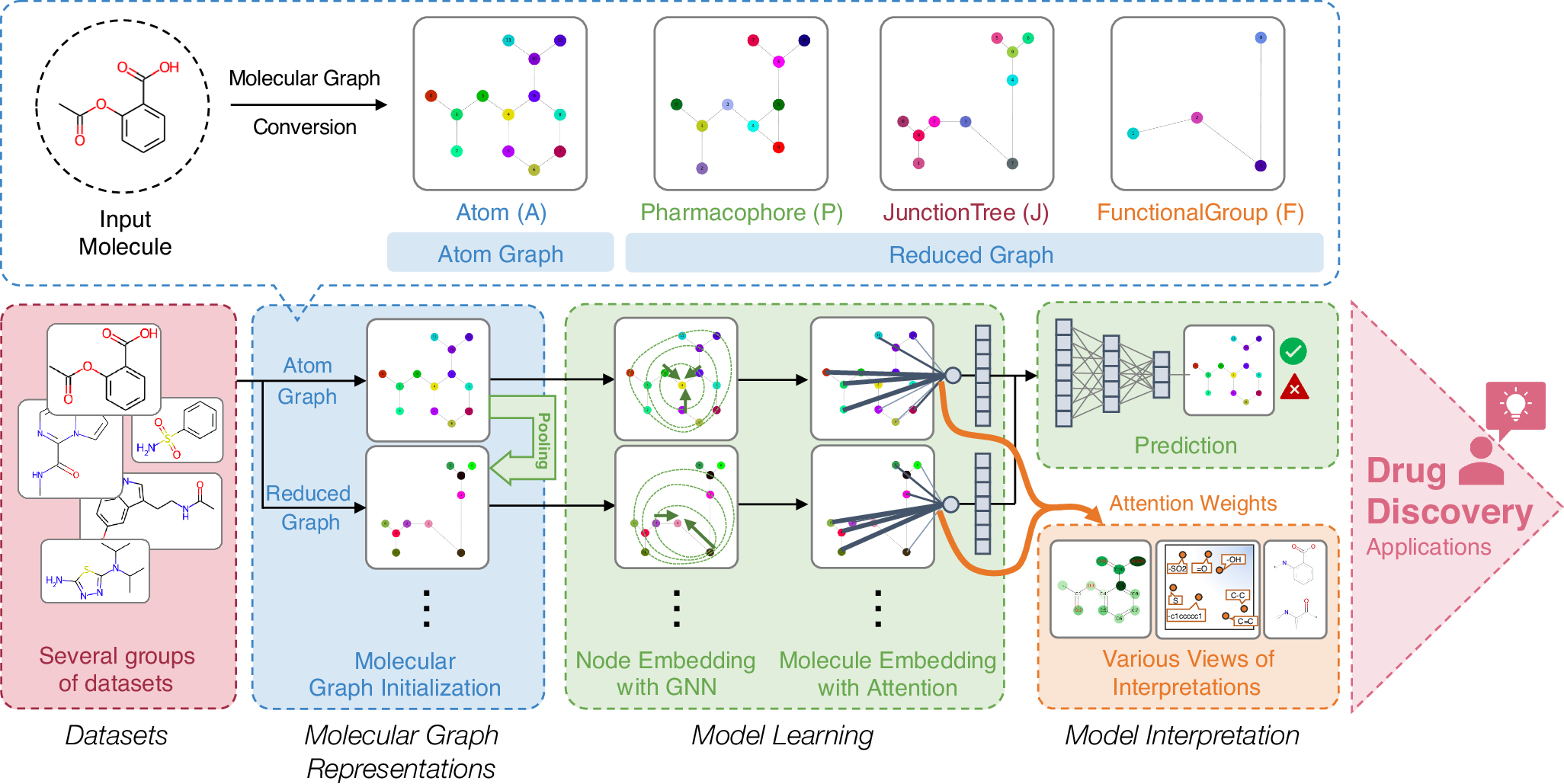
Enhancing property and activity prediction and interpretation using multiple molecular graph representations with MMGX.
Communications Chemistry, 7: 74, 2024. doi: 10.1038/s42004-024-01155-w
 Journal website | PubMed | GitHub
Journal website | PubMed | GitHub
- Ohue M, Sasayama K, Takata M.
Mathematical Modeling and Problem Solving: From Fundamentals to Applications.
The Journal of Supercomputing, 2024. doi: 10.1007/s11227-024-06007-x
 Journal website
Journal website
- Furui K, Ohue M.
FastLomap: Faster Lead Optimization Mapper Algorithm for Large-Scale Relative Free Energy Perturbation.
The Journal of Supercomputing, 2024. doi: 10.1007/s11227-024-06006-y
 Journal website | GitHub
Journal website | GitHub
- Ueki T, Ohue M.
Antibody Complementarity-Determining Region Design using AlphaFold2 and DDG Predictor.
The Journal of Supercomputing, 2024. doi: 10.1007/s11227-023-05887-9
 Journal website | GitHub
Journal website | GitHub
- Hu W, Ohue M.
SpatialPPI: three-dimensional space protein-protein interaction prediction with AlphaFold Multimer.
Computational and Structural Biotechnology Journal, 23: 1214-1225, 2024. doi: 10.1016/j.csbj.2024.03.009
 Journal website | PubMed | bioRxiv | GitHub
Journal website | PubMed | bioRxiv | GitHub
- Ochiai T, Inukai T, Akiyama M, Furui K, Ohue M, Matsumori N, Inuki S, Uesugi M, Sunazuka T, Kikuchi K, Kakeya H, Sakakibara Y.
Variational autoencoder-based chemical latent space for large molecular structures with 3D complexity.
Communications Chemistry, 6: 249, 2023. doi: 10.1038/s42004-023-01054-6
 Journal website | ChemRxiv | PubMed | GitHub
Journal website | ChemRxiv | PubMed | GitHub
- 大上雅史.
AlphaFoldによる高精度なタンパク質立体構造予測と創薬への応用.
PHARM STAGE, 23: 8, 24-28, 2023.
Journal website - Murakumo K, Yoshikawa N, Rikimaru K, Nakamura S, Furui K, Suzuki T, Yamasaki H, Nishigaya Y, Takagi Y, Ohue M.
LLM Drug Discovery Challenge: A Contest as a Feasibility Study on the Utilization of Large Language Models in Medicinal Chemistry.
In Proceedings of AI for Accelerated Materials Design (AI4Mat) NeurIPS 2023 Workshop, 2023.
 OpenReview.net
OpenReview.net
- Ohue M.
MEGADOCK-on-Colab: an easy-to-use protein-protein docking tool on Google Colaboratory.
BMC Research Notes, 16(1): 229, 2023. doi: 10.1186/s13104-023-06505-w
 Journal website | Jxiv | PubMed | GitHub
Journal website | Jxiv | PubMed | GitHub
- Kosugi T, Ohue M.
Design of cyclic peptides targeting protein–protein interactions using AlphaFold.
International Journal of Molecular Sciences, 24(17): 13257, 2023. doi: 10.3390/ijms241713257
 Journal website | bioRxiv | PubMed | GitHub
Journal website | bioRxiv | PubMed | GitHub
- 大上雅史.
AlphaFoldによるタンパク質立体構造予測(実践編).
生物工学会誌 2023年8月号, 101(8): 443-446, 2023. doi: 10.34565/seibutsukogaku.101.8_443
 Journal website
Journal website - Arai M, Makita Y, Saito S, Suzuki S, Narushima Y, Izawa N, Tanaka Y, Furui K, Ohue M, Ishibashi M.
The Indole Rocaglamide Induces S and G2/M Phase Cell Cycle Arrest in Small Cell Lung Cancer Cells Through ASCL1 Translation Inhibition.
SSRN preprint, 2023. doi: 10.2139/ssrn.4493242
 Cell Press Sneak Peak
Cell Press Sneak Peak
- Ohue M, Kojima Y, Kosugi T.
Generating potential protein-protein interaction inhibitor molecules based on physicochemical properties.
Molecules, 28(15): 5652, 2023. doi: 10.3390/molecules28155652
 Journal website | Preprints.org | PubMed | GitHub
Journal website | Preprints.org | PubMed | GitHub
- 大上雅史.
AIによって変わる生命科学.
現代化学 2023年8月号, 28-30, 東京化学同人, 2023.
Book website - Andreani J, Jiménez-García B, Ohue M.
Web Tools for Modeling and Analysis of Biomolecular Interactions Volume II.
Frontiers in Molecular Biosciences, 10: 1190855, 2023. doi: 10.3389/fmolb.2023.1190855
 Journal website | PubMed
Journal website | PubMed - 大上雅史.
PPI(タンパク質間相互作用)を標的とするドラッグデザイン.
タンパク質の構造解析手法とIn silicoスクリーニングへの応用事例, 402-410, 技術情報協会, 2023.
Book website - Kengkanna A, Ohue M.
Enhancing Model Learning and Interpretation Using Multiple Molecular Graph Representations for Compound Property and Activity Prediction.
In Proceedings of The 20th IEEE International Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB 2023), 8 pages, 2023. doi: 10.1109/CIBCB56990.2023.10264879
 Proceedings website | * Download | arXiv | Presentation video
Proceedings website | * Download | arXiv | Presentation video
* © 2023 IEEE. Personal use of this material is permitted. Permission from IEEE must be obtained for all other uses, in any current or future media, including reprinting/republishing this material for advertising or promotional purposes, creating new collective works, for resale or redistribution to servers or lists, or reuse of any copyrighted component of this work in other works. - Ueki T, Ohue M.
Antibody Complementarity-Determining Region Sequence Design using AlphaFold2 and Binding Affinity Prediction Model.
In Proceedings of The 29th International Conference on Parallel & Distributed Processing Techniques and Applications (PDPTA’23), 2133-2139, 2023. doi: 10.1109/CSCE60160.2023.00350
 Proceedings website | Download | bioRxiv
Proceedings website | Download | bioRxiv
- Furui K, Ohue M.
Faster Lead Optimization Mapper Algorithm for Large-Scale Relative Free Energy Perturbation.
In Proceedings of The 29th International Conference on Parallel & Distributed Processing Techniques and Applications (PDPTA’23), 2126-2132, 2023. doi: 10.1109/CSCE60160.2023.00349
 Proceedings website | Download | arXiv | GitHub
Proceedings website | Download | arXiv | GitHub
- Li J, Yanagisawa K, Sugita M, Fujie T, Ohue M, Akiyama Y.
CycPeptMPDB: A Comprehensive Database of Membrane Permeability of Cyclic Peptides.
Journal of Chemical Information and Modeling, 63(7): 2240-2250, 2023. doi: 10.1021/acs.jcim.2c01573
 Journal website | PubMed | Database
Journal website | PubMed | Database - 大上雅史.
中分子ペプチド創薬のインフォマティクス.
実験医学, 2023年1月号, 41(1): 20-26, 羊土社, 2023. doi: 10.18958/7173-00001-0000342-00
Book website - Sugita M, Fujie T, Yanagisawa K, Ohue M, Akiyama Y.
Lipid composition is critical for accurate membrane permeability prediction of cyclic peptides by molecular dynamics simulations.
Journal of Chemical Information and Modeling, 62(18): 4549-4560, 2022. doi: 10.1021/acs.jcim.2c00931
 Journal website | PubMed
Journal website | PubMed - Yanagisawa K, Kubota R, Yoshikawa Y, Ohue M, Akiyama Y.
Effective Protein-Ligand Docking Strategy via Fragment Reuse and a Proof-of-Concept Implementation.
ACS Omega, 7(34): 30265-30274, 2022. doi: 10.1021/acsomega.2c03470
 Journal website | PubMed | GitHub
Journal website | PubMed | GitHub - Kosugi T, Ohue M.
Solubility-aware protein binding peptide design using AlphaFold.
Biomedicines, 10(7): 1626, 2022. doi: 10.3390/biomedicines10071626
 Journal website | PubMed | bioRxiv | GitHub | 日本語の解説
Journal website | PubMed | bioRxiv | GitHub | 日本語の解説 - Furui K, Ohue M.
Compound virtual screening by learning-to-rank with gradient boosting decision tree and enrichment-based cumulative gain.
In Proceedings of The 19th IEEE International Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB 2022), 7 pages, 2022. doi: 10.1109/CIBCB55180.2022.9863032
 Proceedings website | * Download | arXiv | Presentation video* © 2022 IEEE. Personal use of this material is permitted. Permission from IEEE must be obtained for all other uses, in any current or future media, including reprinting/republishing this material for advertising or promotional purposes, creating new collective works, for resale or redistribution to servers or lists, or reuse of any copyrighted component of this work in other works.
Proceedings website | * Download | arXiv | Presentation video* © 2022 IEEE. Personal use of this material is permitted. Permission from IEEE must be obtained for all other uses, in any current or future media, including reprinting/republishing this material for advertising or promotional purposes, creating new collective works, for resale or redistribution to servers or lists, or reuse of any copyrighted component of this work in other works.
- 大上雅史.
AlphaFold2の登場と創薬への影響.
革新的AI創薬~医療ビッグデータ、人工知能がもたらす創薬研究の未来像~, エヌ・ティー・エス, 164-175, 2022.
Book website - Andreani J, Ohue M, Jiménez-García B.
Web Tools for Modeling and Analysis of Biomolecular Interactions.
Frontiers in Molecular Biosciences, 9:875859, 2022. doi: 10.3389/fmolb.2022.875859
 Journal website | PubMed
Journal website | PubMed - 大上雅史.
AlphaFoldのタンパク質立体構造予測の性能.
実験医学, 2022年2月号, 40(2): 427-430, 羊土社, 2022. doi: 10.18958/6977-00002-0000038-00
Book website - 大上雅史.
AlphaFold利用のすすめ.
実験医学, 2022年2月号, 40(2): 433-438, 羊土社, 2022. doi: 10.18958/6977-00002-0000040-00
Book website - Li J, Yanagisawa K, Yoshikawa Y, Ohue M, Akiyama Y.
Plasma protein binding prediction focusing on residue-level features and circularity of cyclic peptides by deep learning.
Bioinformatics, 38(4):1110-1117, 2022. doi: 10.1093/bioinformatics/btab726
 Journal website | PubMed | GitHub
Journal website | PubMed | GitHub - Kosugi T, Ohue M.
Quantitative estimate index for early-stage screening of compounds targeting protein-protein interactions.
International Journal of Molecular Sciences, 22(20): 10925, 2021. doi: 10.3390/ijms222010925
 Journal website | PubMed | GitHub | 日本語の解説
Journal website | PubMed | GitHub | 日本語の解説
- Takabatake K, Izawa K, Akikawa M, Yanagisawa K, Ohue M, Akiyama Y.
Improved large-scale homology search by two-step seed search using multiple reduced amino acid alphabets.
Genes, 12(9): 1455, 2021. doi: 10.3390/genes12091455
 Journal website | PubMed | GitHub
Journal website | PubMed | GitHub - Sugita M, Sugiyama S, Fujie T, Yoshikawa Y, Yanagisawa K, Ohue M, Akiyama Y.
Large-scale membrane permeability prediction of cyclic peptides crossing a lipid bilayer based on enhanced sampling molecular dynamics simulations.
Journal of Chemical Information and Modeling, 61(7): 3681-3695, 2021. doi: 10.1021/acs.jcim.1c00380
 Journal website | PubMed
Journal website | PubMed - Izawa K, Okamoto-Shibayama K, Kita D, Tomita S, Saito A, Ishida T, Ohue M, Akiyama Y, Ishihara K.
Taxonomic and gene category analyses of subgingival plaques from a group of Japanese individuals with and without periodontitis.
International Journal of Molecular Sciences, 22(10): 5298, 2021. doi:10.3390/ijms22105298
 Journal website | PubMed
Journal website | PubMed - Ohue M, Aoyama K, Akiyama Y.
High-performance cloud computing for exhaustive protein-protein docking.
In Proceedings of The 26th International Conference on Parallel & Distributed Processing Techniques and Applications (PDPTA’20), Advances in Parallel & Distributed Processing and Applications, 737-746, 2021. doi:10.1007/978-3-030-69984-0_53
 Proceedings website | arXiv
Proceedings website | arXiv - Ohue M, Akiyama Y.
MEGADOCK-GUI: a GUI-based complete cross-docking tool for exploring protein-protein interactions.
In Proceedings of The 27th International Conference on Parallel & Distributed Processing Techniques and Applications (PDPTA’21), Advances in Parallel & Distributed Processing and Applications. (accepted)
arXiv, Preprint, 2105.03617 [q-bio.BM], 2021.
 Proceedings website | arXiv
Proceedings website | arXiv - Ohue M, Watanabe H, Akiyama Y.
MEGADOCK-Web-Mito: human mitochondrial protein-protein interaction prediction database.
In Proceedings of The 27th International Conference on Parallel & Distributed Processing Techniques and Applications (PDPTA’21), Advances in Parallel & Distributed Processing and Applications. (accepted)
arXiv, Preprint, 2105.00445 [q-bio.BM], 2021.
 Proceedings website | arXiv
Proceedings website | arXiv - Isawa K, Yanagisawa K, Ohue M, Akiyama Y.
Antisense oligonucleotide activity analysis based on opening and binding energies to targets.
In Proceedings of The 27th International Conference on Parallel & Distributed Processing Techniques and Applications (PDPTA’21), Advances in Parallel & Distributed Processing and Applications. (accepted)
Proceedings website - Sugita S, Ohue M.
Drug-target affinity prediction using applicability domain based on data density.
In Proceedings of The 18th IEEE International Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB 2021), 6 pages, 2021. doi:10.1109/CIBCB49929.2021.9562808
 Proceedings website | * Download | ChemRxiv | Presentation video
Proceedings website | * Download | ChemRxiv | Presentation video
* © 2021 IEEE. Personal use of this material is permitted. Permission from IEEE must be obtained for all other uses, in any current or future media, including reprinting/republishing this material for advertising or promotional purposes, creating new collective works, for resale or redistribution to servers or lists, or reuse of any copyrighted component of this work in other works. - Kosugi T, Ohue M.
Quantitative estimate of protein-protein interaction targeting drug-likeness.
In Proceedings of The 18th IEEE International Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB 2021), 8 pages, 2021. doi:10.1109/CIBCB49929.2021.9562931
 Proceedings website | * Download | ChemRxiv | Presentation video
Proceedings website | * Download | ChemRxiv | Presentation video
* © 2021 IEEE. Personal use of this material is permitted. Permission from IEEE must be obtained for all other uses, in any current or future media, including reprinting/republishing this material for advertising or promotional purposes, creating new collective works, for resale or redistribution to servers or lists, or reuse of any copyrighted component of this work in other works. - Ohue M.
Re-ranking of computational protein–peptide docking solutions with amino acid profiles of rigid-body docking results.
In Proceedings of The 21st International Conference on Bioinformatics & Computational Biology (BIOCOMP’20), Advances in Computer Vision and Computational Biology, 749-758, 2021. doi:10.1007/978-3-030-71051-4_58
 Proceedings website | bioRxiv
Proceedings website | bioRxiv - Ito S, Senoo A, Nagatoishi S, Ohue M, Yamamoto M, Tsumoto K, Wakui N.
Structural basis for binding mechanism of human serum albumin complexed with cyclic peptide dalbavancin.
Journal of Medicinal Chemistry, 63(22): 14045–14053, 2020. doi:10.1021/acs.jmedchem.0c01578
 Journal website | bioRxiv | PubMed
Journal website | bioRxiv | PubMed - Launay G†, Ohue M†*, Santero JP, Matsuzaki M, Hilpert C, Uchikoga N, Hayashi T, Martin J*.
Evaluation of CONSRANK-like scoring functions for rescoring ensembles of protein-protein docking poses.
Frontiers in Molecular Biosciences, 7:559005, 2020. doi:10.3389/fmolb.2020.559005
 Journal website | bioRxiv | PubMed
Journal website | bioRxiv | PubMed
† Co-first authors, * Co-corresponding authors - 大上雅史.
構造情報に基づくタンパク質間相互作用の計算予測.
ファインケミカル, 2020年10月号, 49(10): 25-31, シーエムシー出版, 2020.
Book website - Aoyama K, Kakuta M, Matsuzaki Y, Ishida T, Ohue M, Akiyama Y.
Development of computational pipeline software for genome/exome analysis on the K computer.
Supercomputing Frontiers and Innovations, 7(1): 37-54, 2020. doi:10.14529/jsfi200102
 Journal website
Journal website - Aoyama K, Watanabe H, Ohue M, Akiyama Y.
Multiple HPC environments-aware container image configuration workflow for large-scale all-to-all protein-protein docking calculations.
In Proceedings of the 6th Asian Conference on Supercomputing Frontiers (SCFA2020), Lecture Notes in Computer Science, 12082: 23-39, 2020. doi:10.1007/978-3-030-48842-0_2
 Proceedings website
Proceedings website - Matsuno S, Ohue M, Akiyama Y.
Multidomain protein structure prediction using information about residues interacting on multimeric protein interfaces.
Biophysics and Physicobiology, 17: 2-13, 2020. doi:10.2142/biophysico.BSJ-2019050
 Journal website | PubMed
Journal website | PubMed - Chiba S, Ohue M, Gryniukova A, Borysko P, Zozulya S, Yasuo N, Yoshino R, Ikeda K, Shin WH, Kihara D, Iwadate M, Umeyama H, Ichikawa T, Teramoto R, Hsin KY, Gupta V, Kitano H, Sakamoto M, Higuchi A, Miura N, Yura K, Mochizuki M, Ramakrishnan C, Thangakani AM, Velmurugan D, Gromiha MM, Nakane I, Uchida N, Hakariya H, Tan M, Nakamura HK, Suzuki SD, Ito T, Kawatani M, Kudoh K, Takashina S, Yamamoto KZ, Moriwaki Y, Oda K, Kobayashi D, Okuno T, Minami S, Chikenji G, Prathipati P, Nagao C, Mohsen A, Ito M, Mizuguchi K, Honma T, Ishida T, Hirokawa T, Akiyama Y, Sekijima M.
A prospective compound screening contest identified broader inhibitors for Sirtuin 1.
Scientific Reports, 9: 19585, 2019. doi:10.1038/s41598-019-55069-y
 Journal website | PubMed
Journal website | PubMed - 大上雅史, 林孝紀, 秋山泰.
タンパク質間相互作用と複合体構造の予測結果を検索できるウェブサイト「MEGADOCK-Web」.
実験医学, 2019年6月号, 37(9): 1469-1474, 羊土社, 2019.
Book website - Jiang K, Zhang D, Iino T, Kimura R, Nakajima T, Shimizu K, Ohue M, Akiyama Y.
A playful tool for predicting protein-protein docking.
In Proceedings of the 18th International Conference on Mobile and Ubiquitous Multimedia (MUM 2019), Article No. 40, 5 pages, 2019. doi:10.1145/3365610.3368409
 Proceedings website | Download *
Proceedings website | Download *
* © ACM, 2019. This is the author’s version of the work. It is posted here by permission of ACM for your personal use. Not for redistribution. The definitive version was published in MUM2019. - 秋山泰, 大上雅史, 吉川寧, 和久井直樹.
中分子創薬に適した特性を有する環状ペプチドのインシリコ設計.
ペプチド創薬の最前線(木曽良明監修), 70-78, シーエムシー出版, 2019.
Book website - Ohue M, Suzuki SD, Akiyama Y.
Learning-to-rank technique based on ignoring meaningless ranking orders between compounds.
Journal of Molecular Graphics and Modelling, 92: 192-200, 2019. doi:10.1016/j.jmgm.2019.07.009
 Journal website | PubMed | GitHub
Journal website | PubMed | GitHub - Ban T, Ohue M, Akiyama Y.
NRLMFβ: beta-distribution-rescored neighborhood regularized logistic matrix factorization for improving performance of drug–target interaction prediction.
Biochemistry and Biophysics Reports, 18: 100615, 2019. doi:10.1016/j.bbrep.2019.01.008
 Journal website | PubMed | GitHub
Journal website | PubMed | GitHub - Mochizuki M, Suzuki SD, Yanagisawa K, Ohue M, Akiyama Y.
QEX: Target-specific druglikeness filter enhances ligand-based virtual screening.
Molecular Diversity, 23(1): 11–18, 2019. doi:10.1007/s11030-018-9842-3
 Journal website | PubMed
Journal website | PubMed - Yamamoto K, Yoshikawa Y, Ohue M, Inuki S, Ohno H, Oishi S.
Synthesis of triazolo- and oxadiazolo-piperazines by gold(I)-catalyzed domino cyclization: application to the design of a mitogen activated protein (MAP) kinase inhibitor.
Organic Letters, 21(2): 373-377, 2019. doi:10.1021/acs.orglett.8b03500
Journal website | PubMed - Ohue M, Yamasawa M, Izawa K, Akiyama Y.
Parallelized pipeline for whole genome shotgun metagenomics with GHOSTZ-GPU and MEGAN.
In Proceedings of the 19th annual IEEE International Conference on Bioinformatics and Bioengineering (IEEE BIBE 2019), 152-156, 2019. doi:10.1109/BIBE.2019.00035
 Download * | Proceedings website | slide
Download * | Proceedings website | slide
* © 2019 IEEE. Personal use of this material is permitted. Permission from IEEE must be obtained for all other uses, in any current or future media, including reprinting/republishing this material for advertising or promotional purposes, creating new collective works, for resale or redistribution to servers or lists, or reuse of any copyrighted component of this work in other works. - Ohue M, Ii R, Yanagisawa K, Akiyama Y.
Molecular activity prediction using graph convolutional deep neural network considering distance on a molecular graph.
In Proceedings of the 2019 International Conference on Parallel and Distributed Processing Techniques & Applications (PDPTA’19), 122-128, 2019.
 Proceedings website | arXiv | slide
Proceedings website | arXiv | slide - Aoyama K, Yamamoto Y, Ohue M, Akiyama Y.
Performance evaluation of MEGADOCK protein-protein interaction prediction system implemented with distributed containers on a cloud computing environment.
In Proceedings of the 2019 International Conference on Parallel and Distributed Processing Techniques & Applications (PDPTA’19), 175-181, 2019.
 Proceedings website
Proceedings website - Tajimi T, Wakui N, Yanagisawa K, Yoshikawa Y, Ohue M, Akiyama Y.
Computational prediction of plasma protein binding of cyclic peptides from small molecule experimental data using sparse modeling techniques.
BMC Bioinformatics, 19(Suppl 19): 527, 2018. doi:10.1186/s12859-018-2529-z
 Journal website | PubMed
Journal website | PubMed - Kami D, Kitani T, Nakamura A, Wakui N, Mizutani R, Ohue M, Kametani F, Akimitsu N, Gojo S.
The DEAD-box RNA-binding protein DDX6 regulates parental RNA decay for cellular reprogramming to pluripotency.
PLoS ONE, 13(10): e0203708, 2018. doi:10.1371/journal.pone.0203708
 Journal website | PubMed
Journal website | PubMed - Yanagisawa K, Komine S, Kubota R, Ohue M, Akiyama Y.
Optimization of memory use of fragment extension-based protein-ligand docking with an original fast minimum cost flow algorithm.
Computational Biology and Chemistry, 74: 399-406, 2018. doi:10.1016/j.compbiolchem.2018.03.013
 Journal website | PubMed | Slide
Journal website | PubMed | Slide - Hayashi T, Matsuzaki Y, Yanagisawa K, Ohue M*, Akiyama Y*.
MEGADOCK-Web: an integrated database of high-throughput structure-based protein-protein interaction predictions.
BMC Bioinformatics, 19(Suppl 4): 62, 2018. doi:10.1186/s12859-018-2073-x
 Journal website | PubMed | Slide
Journal website | PubMed | Slide - Ban T, Ohue M, Akiyama Y.
Multiple grid arrangement improves ligand docking with unknown binding sites: Application to the inverse docking problem.
Computational Biology and Chemistry, 73: 139-146, 2018. doi:10.1016/j.compbiolchem.2018.02.008
 Journal website | PubMed | Software
Journal website | PubMed | Software - Suzuki SD, Ohue M, Akiyama Y.
PKRank: A novel learning-to-rank method for ligand-based virtual screening using pairwise kernel and RankSVM.
Artificial Life and Robotics, 23(2): 205-212, 2018. doi:10.1007/s10015-017-0416-8
 Journal website
Journal website - Wakui N, Yoshino R, Yasuo N, Ohue M, Sekijima M.
Exploring the selectivity of inhibitor complexes with Bcl-2 and Bcl-XL: a molecular dynamics simulation approach.
Journal of Molecular Graphics and Modelling, 79: 166-174, 2018. doi:10.1016/j.jmgm.2017.11.011
 Journal website | PubMed
Journal website | PubMed - Yanagisawa K, Komine S, Suzuki SD, Ohue M, Ishida T, Akiyama Y.
Spresso: An ultrafast compound pre-screening method based on compound decomposition.
Bioinformatics, 33(23): 3836-3843, 2017. doi:10.1093/bioinformatics/btx178
 Journal website | PubMed | Software
Journal website | PubMed | Software - Matsuzaki Y, Uchikoga N, Ohue M, Akiyama Y.
Rigid-docking approaches to explore protein-protein interaction space.
Advances in Biochemical Engineering/Biotechnology, 160: 33-55, 2017. doi:10.1007/10_2016_41
Journal website | PubMed - Suzuki S, Ishida T, Ohue M, Kakuta M, Akiyama Y.
GHOSTX: a fast sequence homology search tool for functional annotation of metagenomic data.
Methods in Molecular Biology, 1611: 15-25, 2017. doi:10.1007/978-1-4939-7015-5_2
Journal website | PubMed - Ohue M, Yamazaki T, Ban T, Akiyama Y.
Link mining for kernel-based compound-protein interaction predictions using a chemogenomics approach.
In the Thirteenth International Conference on Intelligent Computing (ICIC2017), Lecture Notes in Computer Science, 10362: 549-558, 2017. doi:10.1007/978-3-319-63312-1_48
 Proceedings website | arXiv | slide
Proceedings website | arXiv | slide - Ban T, Ohue M, Akiyama Y.
Efficient hyperparameter optimization by using Bayesian optimization for drug-target interaction prediction.
In Proceedings of the 7th IEEE International Conference on Computational Advances in Bio and Medical Sciences (IEEE ICCABS 2017), 8 pages, 2017. doi:10.1109/ICCABS.2017.8114299
 Proceedings website | Download * | GitHub
Proceedings website | Download * | GitHub - Suzuki SD, Ohue M, Akiyama Y.
Learning-to-rank based compound virtual screening by using pairwise kernel with multiple heterogeneous experimental data.
In Proceedings of 22nd International Symposium on Artificial Life and Robotics (AROB 22nd 2017), 114-119, 2017.
Proceedings website | slide - Uchikoga N, Matsuzaki Y, Ohue M, Akiyama Y.
Specificity of broad protein interaction surfaces for proteins with multiple binding partners.
Biophysics and Physicobiology, 13: 105-115, 2016. doi:10.2142/biophysico.13.0_105
 Journal website | PubMed
Journal website | PubMed - Yanagisawa K, Komine S, Suzuki SD, Ohue M, Ishida T, Akiyama Y.
ESPRESSO: An ultrafast compound pre-screening method based on compound decomposition.
In Proceedings of The 27th International Conference on Genome Informatics (GIW 2016), 2016.
slide - 大上雅史.
学振申請書の書き方とコツ DC/PD獲得を目指す若者へ.
192 pages, 講談社, 2016.
Book website - Shimoda T, Suzuki S, Ohue M, Ishida T, Akiyama Y.
Protein-protein docking on hardware accelerators: comparison of GPU and MIC architectures.
BMC Systems Biology, 9(Suppl 1): S6, 2015. doi:10.1186/1752-0509-9-S1-S6
 Journal website | PubMed | Slide
Journal website | PubMed | Slide - 大上雅史.
これだけ!生化学(第8章 核酸の生化学).
生化学若い研究者の会著, 稲垣賢二監修, 秀和システム, 2014.
Book website - Ohue M, Shimoda T, Suzuki S, Matsuzaki Y, Ishida T, Akiyama Y.
MEGADOCK 4.0: an ultra-high-performance protein-protein docking software for heterogeneous supercomputers.
Bioinformatics, 30(22): 3281-3283, 2014. doi:10.1093/bioinformatics/btu532
 Journal website | PubMed | GitHub
Journal website | PubMed | GitHub - Ohue M, Matsuzaki Y, Uchikoga N, Ishida T, Akiyama Y.
MEGADOCK: An all-to-all protein-protein interaction prediction system using tertiary structure data.
Protein and Peptide Letters, 21(8): 766-778, 2014. doi:10.2174/09298665113209990050
 Journal website | PubMed | GitHub
Journal website | PubMed | GitHub - Matsuzaki Y, Ohue M, Uchikoga N, Akiyama Y.
Protein-protein interaction network prediction by using rigid-body docking tools: application to bacterial chemotaxis.
Protein and Peptide Letters, 21(8): 790-798, 2014. doi:10.2174/09298665113209990066
 Journal website | PubMed
Journal website | PubMed - Ohue M, Matsuzaki Y, Shimoda T, Ishida T, Akiyama Y.
Highly precise protein-protein interaction prediction based on consensus between template-based and de novo docking methods.
BMC Proceedings, 7(Suppl 7): S6, 2013. doi:10.1186/1753-6561-7-S7-S6
 Journal website | PubMed
Journal website | PubMed - Matsuzaki Y, Uchikoga N, Ohue M, Shimoda T, Sato T, Ishida T, Akiyama Y.
MEGADOCK 3.0: a high-performance protein-protein interaction prediction software using hybrid parallel computing for petascale supercomputing environments.
Source Code for Biology and Medicine, 8(1): 18, 2013. doi:10.1186/1751-0473-8-18
 Journal website | PubMed | GitHub
Journal website | PubMed | GitHub - 中嶋悠介, 大上雅史, 越野亮
配列情報に基づくタンパク質間相互作用予測の構造情報付加による高精度化.
FIT2013 第12回情報科学技術フォーラム講演論文集, 第2分冊(RG-001): 63-68, 2013.
 Download*
Download*
* 本論文は一般社団法人 電子情報通信学会および一般社団法人 情報処理学会の著作権規程に基いて公開しているものです. - Uchikoga N, Matsuzaki Y, Ohue M, Hirokawa T, Akiyama Y.
Re-docking scheme for generating near-native protein complexes by assembling residue interaction fingerprints.
PLoS ONE, 8(7): e69365, 2013. doi:10.1371/journal.pone.0069365
 Journal website | PubMed
Journal website | PubMed - Shimoda T, Ishida T, Suzuki S, Ohue M, Akiyama Y.
MEGADOCK-GPU: Acceleration of Protein-Protein Docking Calculation on GPUs.
In Proceedings of the ACM Conference on Bioinformatics, Computational Biology and Biomedicine 2013 (ACM-BCB 2013), 2nd International Workshop on Parallel and Cloud-based Bioinformatics and Biomedicine (ParBio2013), 884-890, 2013. doi:10.1145/2506583.2506693
 Proceedings website | Download * | slide
Proceedings website | Download * | slide
* © ACM, 2013. This is the author’s version of the work. It is posted here by permission of ACM for your personal use. Not for redistribution. The definitive version was published in BCB’13. - Ohue M, Matsuzaki Y, Shimoda T, Ishida T, Akiyama Y.
Highly precise protein-protein interaction prediction based on consensus between template-based and de novo docking methods.
In Proceedings of Great Lakes Bioinformatics Conference 2013 (GLBIO2013), 100-109, 2013.
 Download | slide
Download | slide - Ohue M, Matsuzaki Y, Ishida T, Akiyama Y.
Improvement of the protein-protein docking prediction by introducing a simple hydrophobic interaction model: an application to interaction pathway analysis.
In Proceedings of The 7th IAPR International Conference on Pattern Recognition in Bioinformatics (PRIB2012), Lecture Notes in Computer Science, 7632: 178-187, 2012. doi:10.1007/978-3-642-34123-6_16
Proceedings website | slide - Fleishman SJ, Whitehead TA, Strauch EM, Corn JE, Qin S, Zhou HX, Mitchell JC, Demerdash ON, Takeda-Shitaka M, Terashi G, Moal IH, Li X, Bates PA, Zacharias M, Park H, Ko JS, Lee H, Seok C, Bourquard T, Bernauer J, Poupon A, Azé J, Soner S, Ovali SK, Ozbek P, Tal NB, Haliloglu T, Hwang H, Vreven T, Pierce BG, Weng Z, Pérez-Cano L, Pons C, Fernández-Recio J, Jiang F, Yang F, Gong X, Cao L, Xu X, Liu B, Wang P, Li C, Wang C, Robert CH, Guharoy M, Liu S, Huang Y, Li L, Guo D, Chen Y, Xiao Y, London N, Itzhaki Z, Schueler-Furman O, Inbar Y, Potapov V, Cohen M, Schreiber G, Tsuchiya Y, Kanamori E, Standley DM, Nakamura H, Kinoshita K, Driggers CM, Hall RG, Morgan JL, Hsu VL, Zhan J, Yang Y, Zhou Y, Kastritis PL, Bonvin AM, Zhang W, Camacho CJ, Kilambi KP, Sircar A, Gray JJ, Ohue M, Uchikoga N, Matsuzaki Y, Ishida T, Akiyama Y, Khashan R, Bush S, Fouches D, Tropsha A, Esquivel-Rodríguez J, Kihara D, Stranges PB, Jacak R, Kuhlman B, Huang SY, Zou X, Wodak SJ, Janin J, Baker D.
Community-wide assessment of protein-interface modeling suggests improvements to design methodology.
Journal of Molecular Biology, 414(2): 289-302, 2011. doi: 10.1016/j.jmb.2011.09.031
Journal website | PubMed - Ohue M, Matsuzaki Y, Akiyama Y.
Docking-calculation-based method for predicting protein-RNA interactions.
Genome Informatics, 25(1): 25-39, 2011. doi:10.11234/gi.25.25
 Journal website | PubMed
Journal website | PubMed - 大上雅史, 松崎由理, 松﨑裕介, 佐藤智之, 秋山泰.
MEGADOCK:立体構造情報からの網羅的タンパク質間相互作用予測とそのシステム生物学への応用.
情報処理学会論文誌 数理モデル化と応用, 3(3): 91-106, 2010.
 Journal website
Journal website




* © 2017 IEEE. Personal use of this material is permitted. Permission from IEEE must be obtained for all other uses, in any current or future media, including reprinting/republishing this material for advertising or promotional purposes, creating new collective works, for resale or redistribution to servers or lists, or reuse of any copyrighted component of this work in other works.
